Post 46: An AI that creates proteins from scratch 🎩🪄
Published:
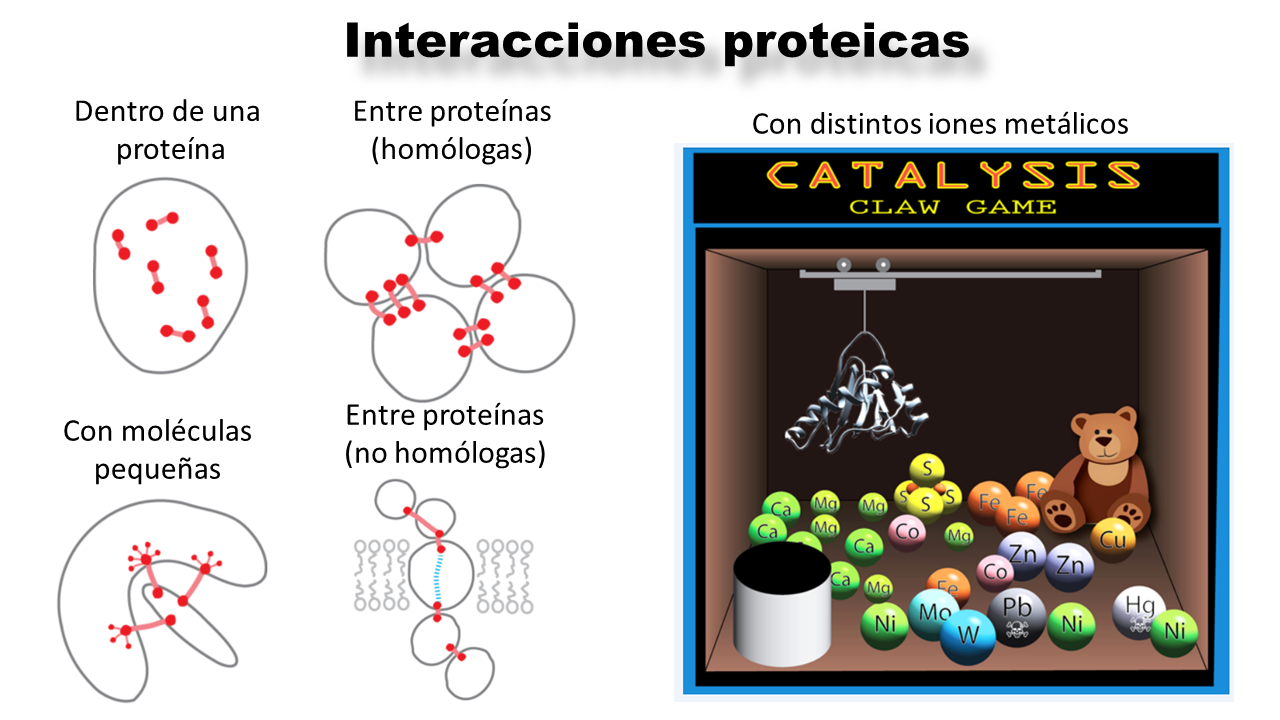
For a protein to function, it must first fold. For this to happen, certain amino acids must come into contact to favor the necessary energetic and geometric conditions. However, proteins rarely act alone; they usually interact with each other, with DNA/RNA, with other small molecules (like antibiotics), and even with metal ions (as in photosynthesis). When a protein interacts in these ways, we call it a protein complex.
AlphaFold2 has revolutionized the science of proteins, but its major weakness is that it cannot predict the chemical bonds that occur in a protein complex (in fact, it also cannot model interactions like disulfide bonds). To try to solve this, in David Baker’s laboratory, they have trained RoseTTAFold All-Atom (RFAA), a new AI capable of creating proteins from scratch.
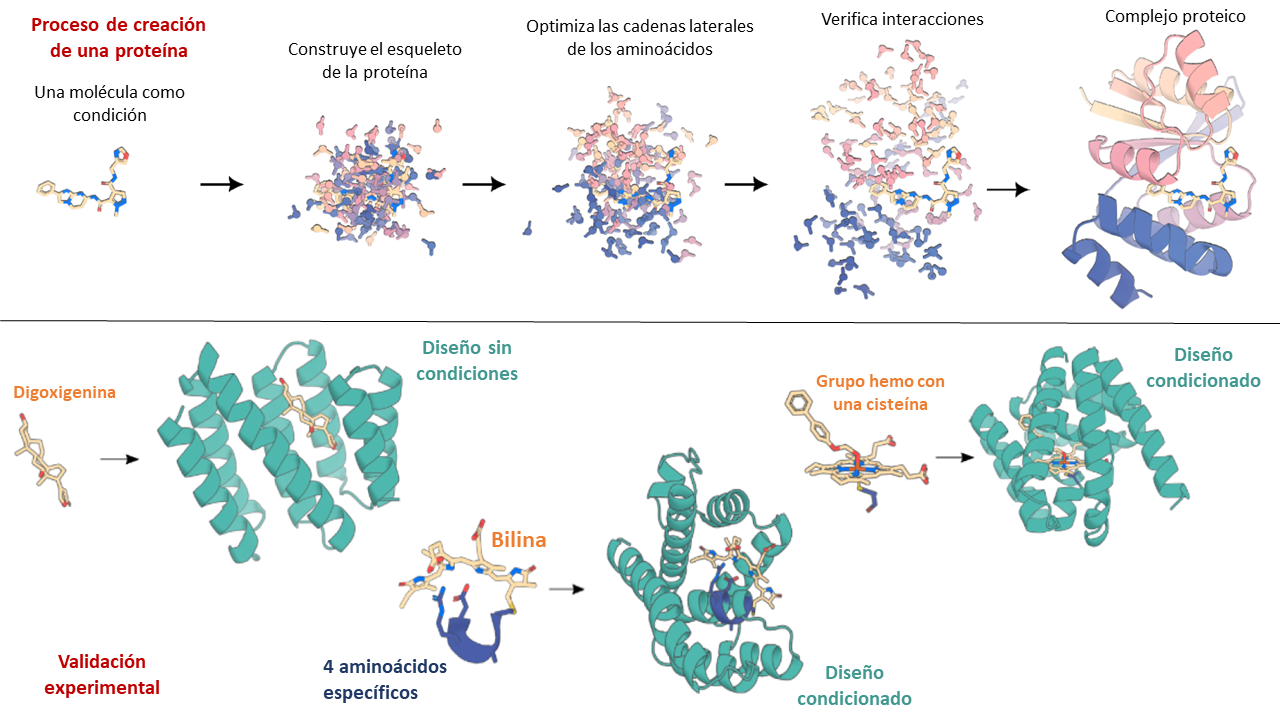
RFAA was trained paying attention to the chemical bonds that are established when proteins interact. To do this, they used many protein complexes, and furthermore, they represented atom by atom the regions that are interacting (and not amino acid by amino acid, as is normally done). With this, they managed to make the AI build a protein that satisfies a specific interaction, such as that between an enzyme and an antibiotic. That is, it is an AI that constructs on a specific template (interaction).
The capabilities of this AI to create new proteins have been demonstrated by experimentally verifying three cases where the proteins manage to fold and fulfill their function. In the first case, they let the AI create a protein from scratch that could bind to a steroid called digoxigenin (used for heart diseases). In the second case, they asked it to create a protein that would bind to a heme group (like that of hemoglobin) around a cysteine. And in the third case, they asked for a protein that would bind through four specific amino acids to pigments called bilins that are involved in photosynthesis
The most interesting thing is that the proteins created by AI do not resemble any protein used in training, which suggests that the AI learned notions of the physics and geometry of protein interactions. However, the authors acknowledge that it is necessary to improve its performance using more training data. And for the moment, they have not released their code, although we can expect it in about 6/8 months when the article is officially published.
refs: