Post 6: Protein language models 💻
Published:
Previously, I wrote about how to think of proteins as languages. Here, I provide more details about one of these protein languages (ESM-1b).
Modeling biology is difficult because it tends to be synergistic, emergent, and context-dependent, rather than additive. Since biological systems are more than the sum of their parts, making useful predictions solely from them is extremely complicated. It gets even worse when you consider that they evolve over time and what we see today is just a tiny fragment of the biodiversity that has existed on Earth (the historical property of biology). So, a model that manages to capture so much information seemed impossible to achieve until recently.
In the field of artificial intelligence, natural language processing is a strategy that has allowed for the “semanticization” of tons of information. The easiest way to understand these models (for example, GPT-J) is by generating coherent text given a base phrase. In biology, this is reflected in many ways, such as learning to group proteins according to their secondary structure (F1).
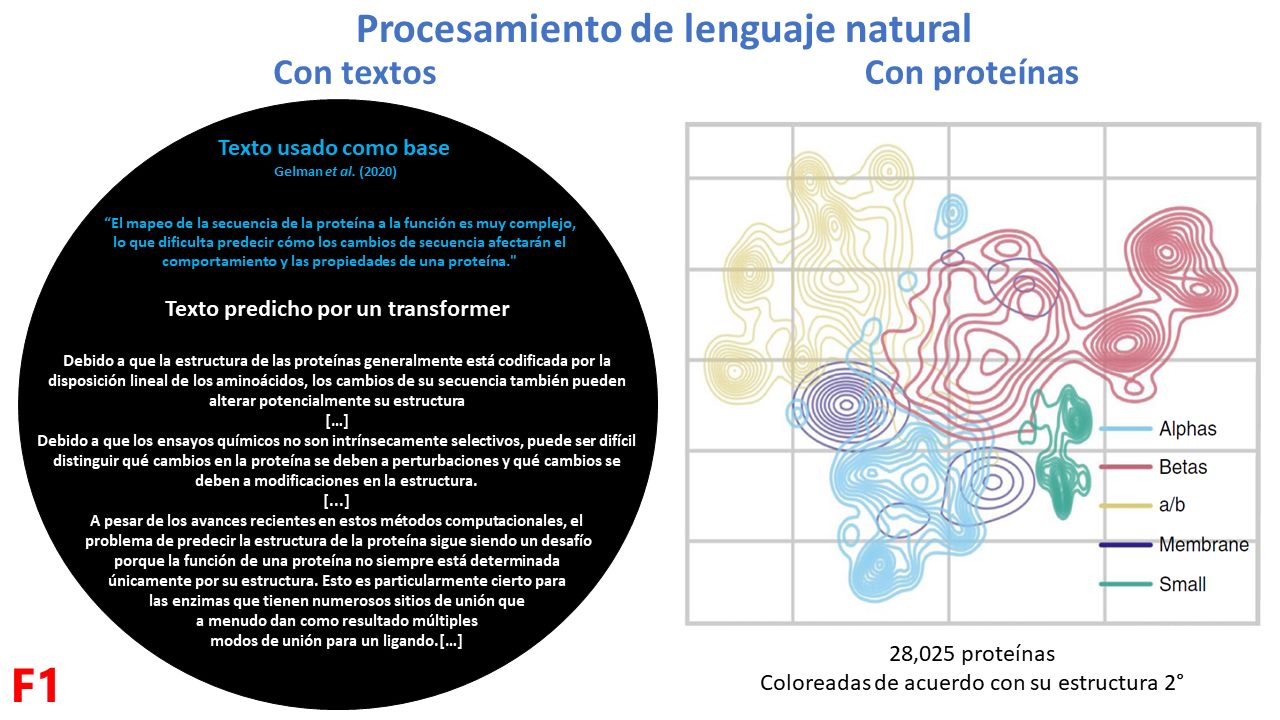
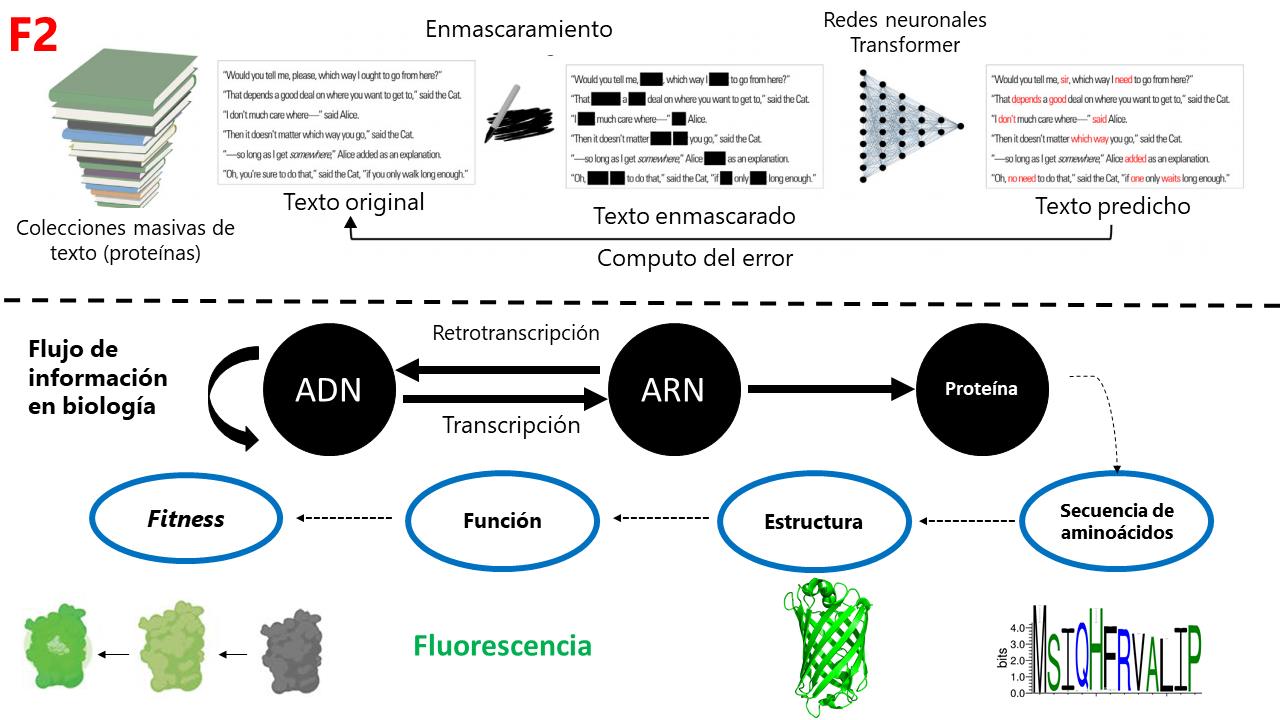
ESM-1b (Evolutionary Scale Modeling) is a model that has learned biology by observing 250 million proteins. Its premise is that the proteins we see today are also “informational fossils” that, having been shaped by evolution, contain “rules” of what is more biophysically probable. ESM-1b learned biology by taking each of the proteins, masking some of its amino acids, and comparing this masked protein against its original version to compute which amino acid combinations are most probable. This allowed it to “distill” emergent properties such as the structure and function of proteins, and even the degree of functionality (or fitness) of proteins (F2).
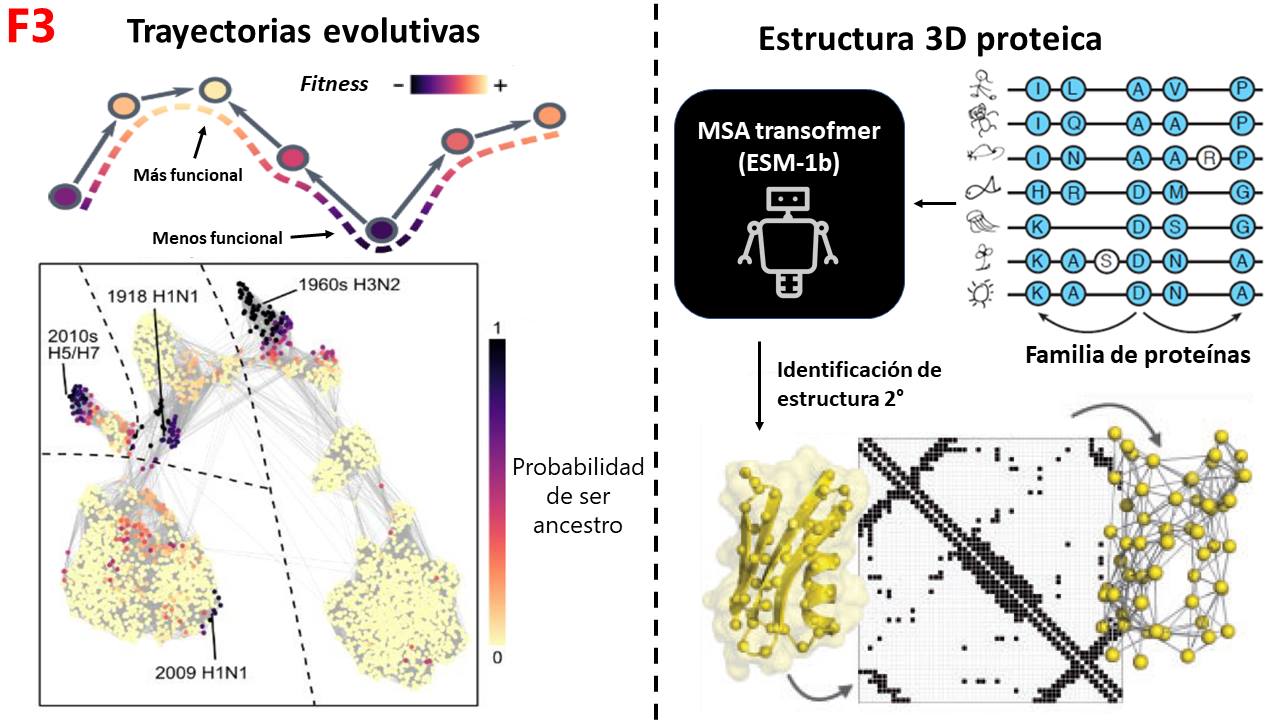
ESM-1b can be used to predict the fitness of proteins and trace evolutionary trajectories from less to more functional; and then, assign which proteins were probably the ancestors of a whole set. If we apply this to the influenza virus, we can see that ESM-1b identifies proteins that originated in pandemics, such as those of the 20th century or the 2009 pandemic, as origin points (ancestors), allowing us to model part of the dynamics of pandemics. ESM-1b can also be used to identify the 3D structure of proteins (MSA-Transformer). For this, specialized training is done with proteins from the same family to learn to distinguish which regions of amino acids are most likely to have certain secondary structures and thus identify the complete 3D folding. In fact, this same strategy is used by AlphaFold2 with a module called Evoformer (F3).
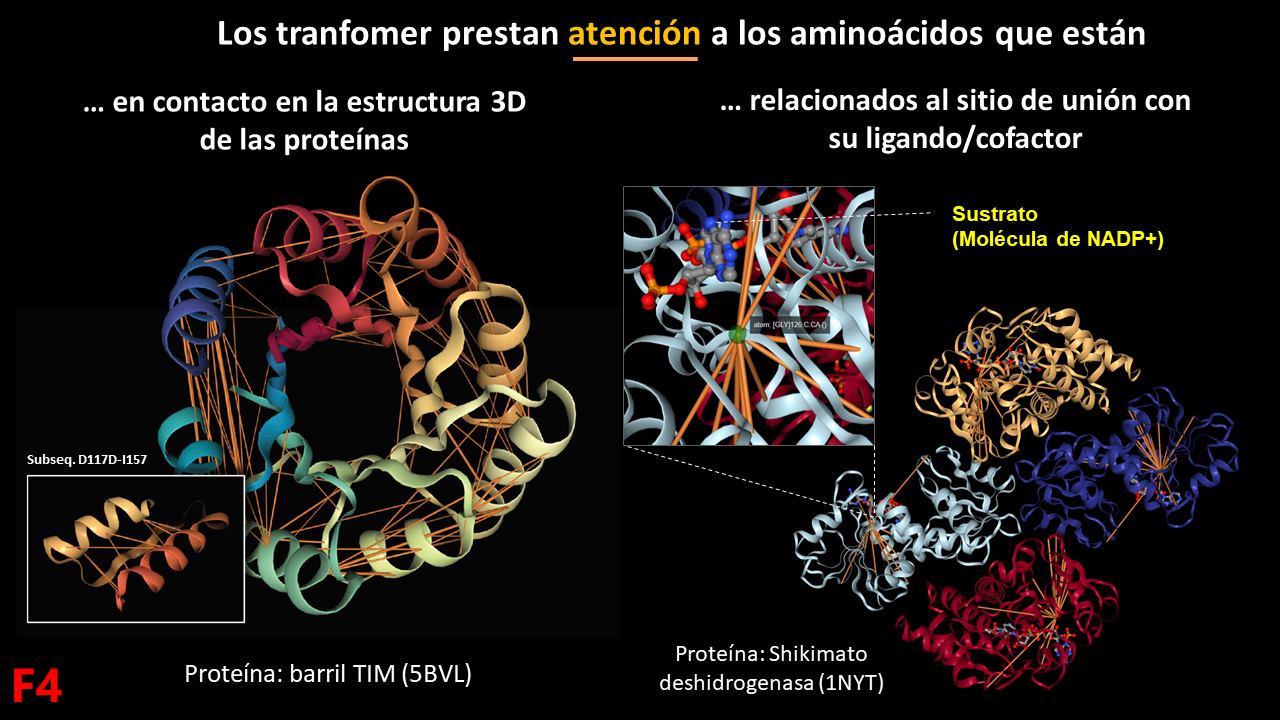
At the heart of ESM-1b is a mathematical operation known as “Attention,” which characterizes a type of neural network known as “Transformers.” Attention allows transformers to identify the most relevant parts of a sequence of information. If we look at it in proteins, the amino acids that have more attention between them are those that are in 3D contact in the protein, and there is even a lot of attention to the amino acids related to the substrate binding site in the case of enzymes. This demonstrates that during the training of ESM-1b with millions of proteins, in each case, it pays attention to the most important amino acids for structure and function.
The applications of ESM-1b and other transformers are just beginning. In fact, there is another model (ProGen) that has learned to synthesize proteins just by telling it the type of protein you want (e.g., a 222 amino acid long hydrolase from Homo sapiens). What awaits us in this decade in the area of natural language processing applied to biology is exciting! Quite the opposite of what climate change holds for us…
I wouldn’t want to go anywhere without my incredible towel.
Refs:
ESM-1b
Protein language models applied to structure prediction
- Transformer protein language models are unsupervised structure learners
- BERTology Meets Biology: Interpreting Attention in Protein Language Models
Protein language models applied to function prediction
- Language models enable zero-shot prediction of the effects of mutations on protein function
- Evolutionary velocity with protein language models
- Evotuning protocols for Transformer-based variant effect prediction on multi-domain proteins
ProGen*
