Post 4: How to think about proteins? 🤔
Published:
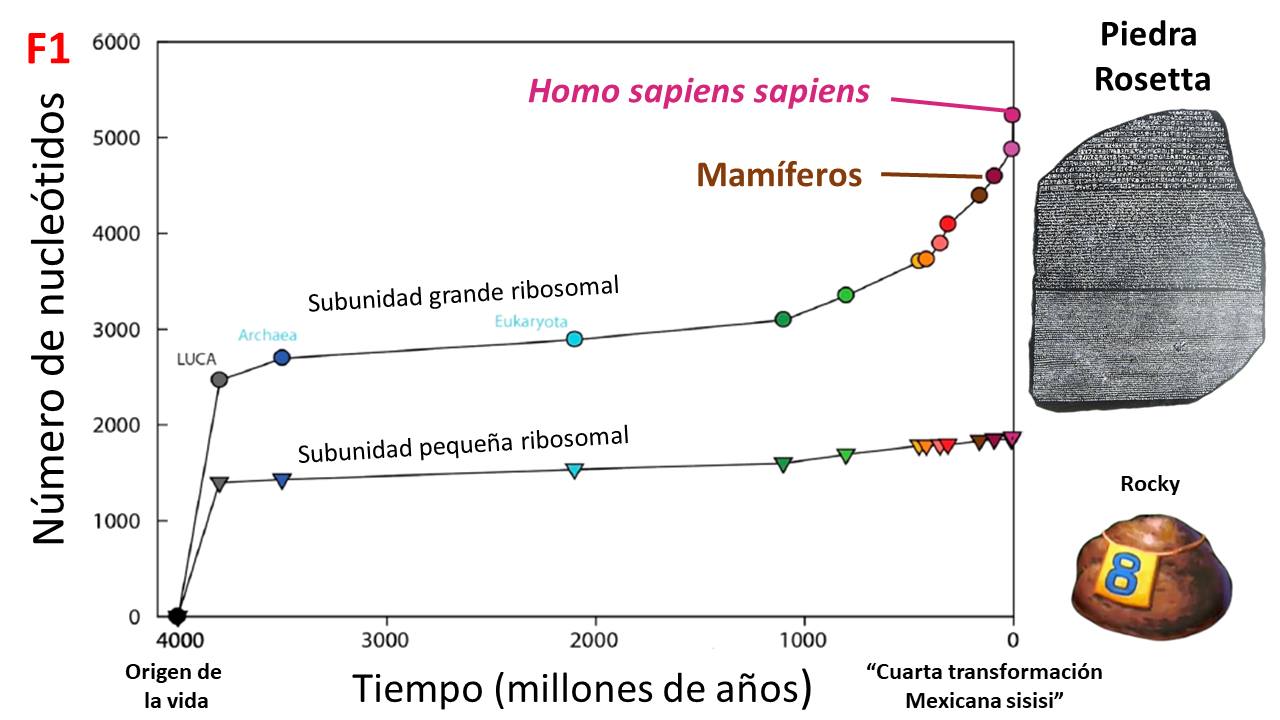
1) Like clocks: some will give you the hour while others will even give you seconds. This information load of proteins is due to the patterns in how they accumulate mutations in their genes, which are reflected in amino acids subject to different evolutionary forces. This idea has helped us to date the origin of life on Earth, for example, using ribosomal proteins (F1)
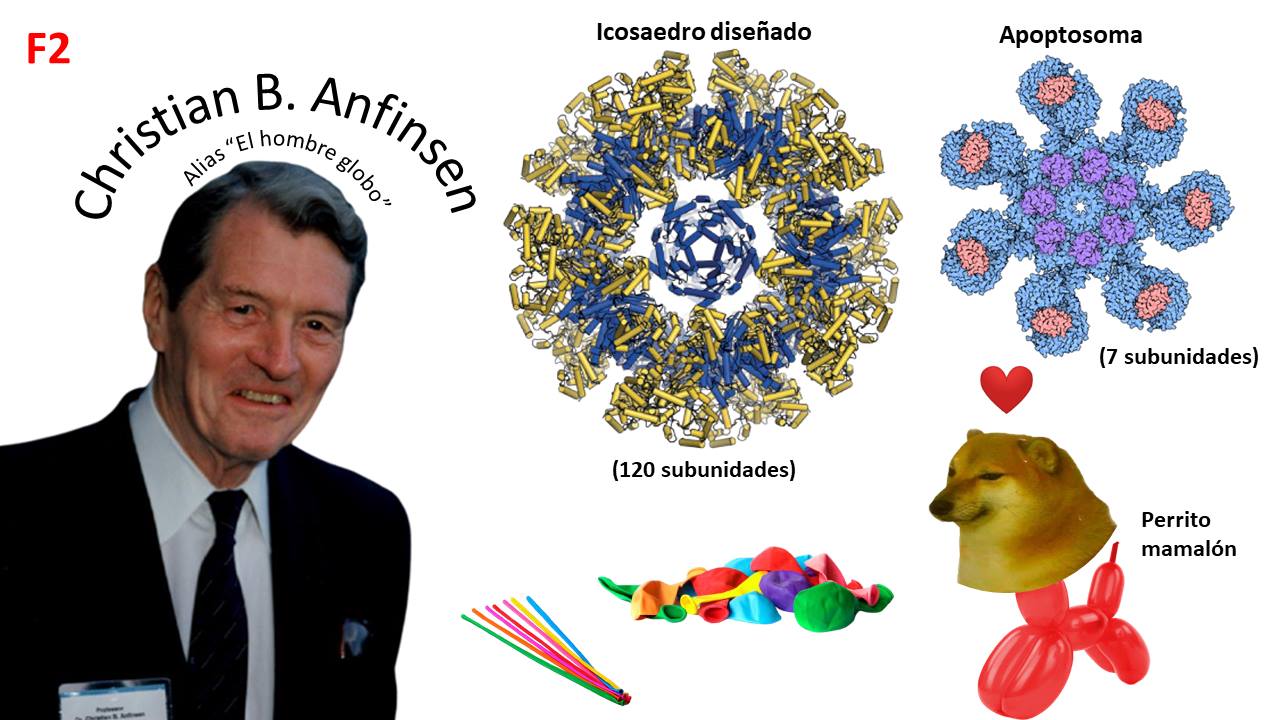
2) Like balloons: some allow for rare shapes like “apoptosomes,” while others are better left untouched because they explode. Just as balloons take shape from their container and air, proteins fold based on the interaction between their amino acids and the thermodynamics that occur in the solvent (water). Christian Anfinsen proposed that all the information for a protein to fold is contained in its amino acids. Today, this idea is reflected in AlphaFold, a set of neural networks that predict the 3D structure of a protein at a good resolution.
3) Like languages: proteins not only have temporal and structural information encoded but also functional. Their amino acids have a semantics. In this sense, they are similar to the Rosetta Stone, which had a message encrypted in three different languages. The functions and how well proteins perform depend on both the identity and 3D position of the amino acids.
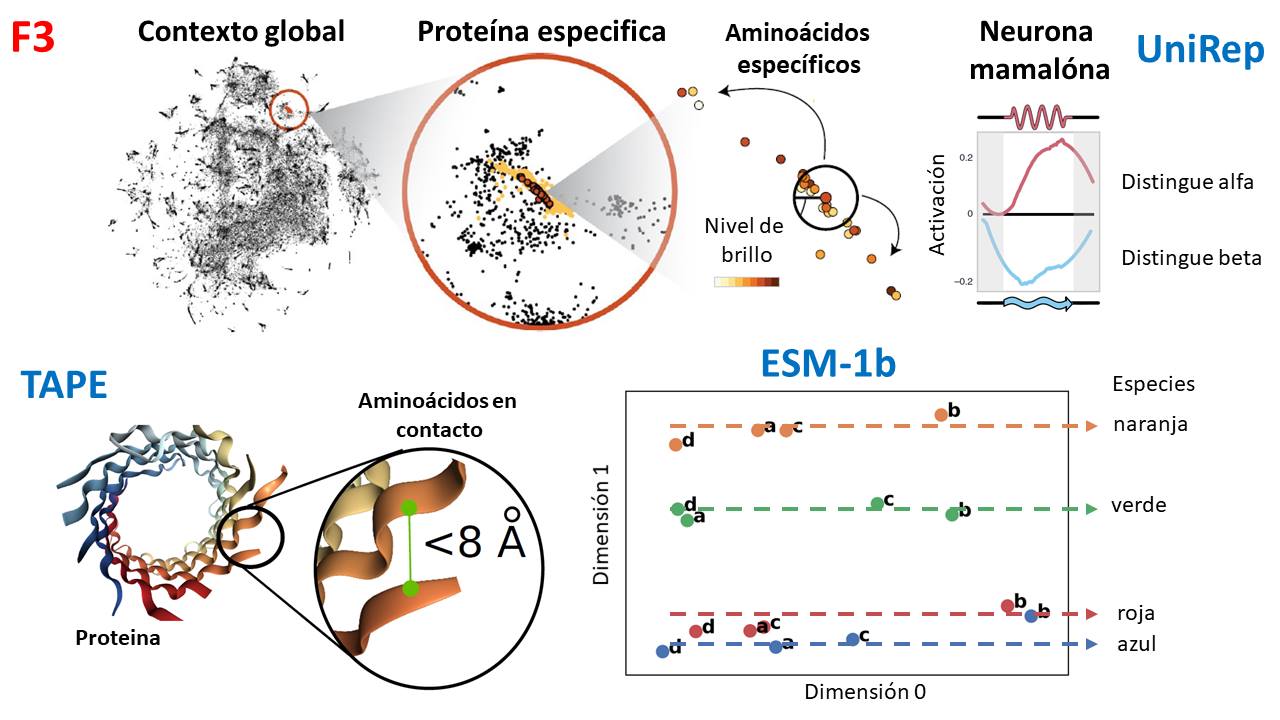
Using neural networks and natural language processing methods (like BERT), we can train models to learn how to encode the semantics of amino acids and then ask these models for information on the evolutionary history, 3D structure, and function of proteins. These models can even create artificial proteins under biophysical limits that one indicates. There are many “neural languages of proteins,” but the following caught my attention (F3):
- “UniRep”: developed at Harvard, trained with 24 million proteins, and found a neuron capable of distinguishing between protein foldings (alpha and beta). It is also possible to fine-tune the model to ask which amino acids would improve a trait of a protein, for example, the brightness of a fluorescent protein.
- “TAPE”: developed at Google and Berkeley, propose a series of metrics to evaluate neural languages of proteins and also train a type of neural networks known as “Transformers” with 31 million proteins, finding that it is capable of predicting how proteins fold to improve their stability.
- “ESM-1b:” developed at Facebook and US universities, is one of the two largest models trained with 250 million proteins and is able to distinguish which type of proteins (orange, green, red, and blue) correspond to which type of species (A, B, C, and D).
The important thing is that these properties were learned ONLY from amino acid sequences; no other type of information was used for training. That is, the networks learned the semantics and emergent aspects of proteins, such as their evolutionary relationships. Which is incredible.
Eat fruits and vegetables.
Refs:
- Proteins as clocks (evolution):
- Proteins as ballons (biophysics):
- Protein as languages:
- Protein language models:
