Post 3: Protein engineering based on artificial intelligence 🤖
Published:
I would like to say that biology has undergone three major analytical transformations:
- A “molecularization” with the characterization of DNA structure.
- A digitalization with massive sequencing technologies.
- A “semanticization” with the massive computation of data.
To provide context to the data, machine learning techniques are used, where neural networks have been the most famous (F1). Within the wide variety of biological data that can be analyzed, the 3D structures of proteins are a rich source of information that can be used to understand their function and improve their capabilities. 
For this purpose, Russ’s laboratory took protein structures and divided them into small pieces that represent the chemical microenvironments of the C, O, N, and S atoms in order to predict regions within the protein that are prone to change. A few years later, Ross’s team would take Russ’s neural network and improve it by adding more detailed information, for example, considering all the H.
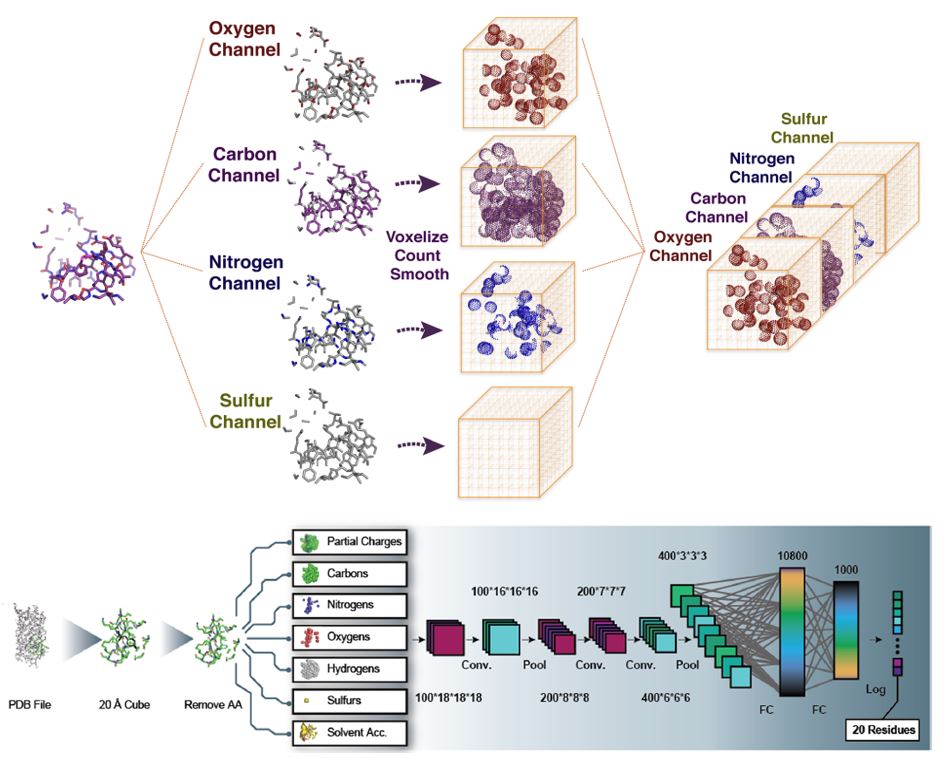
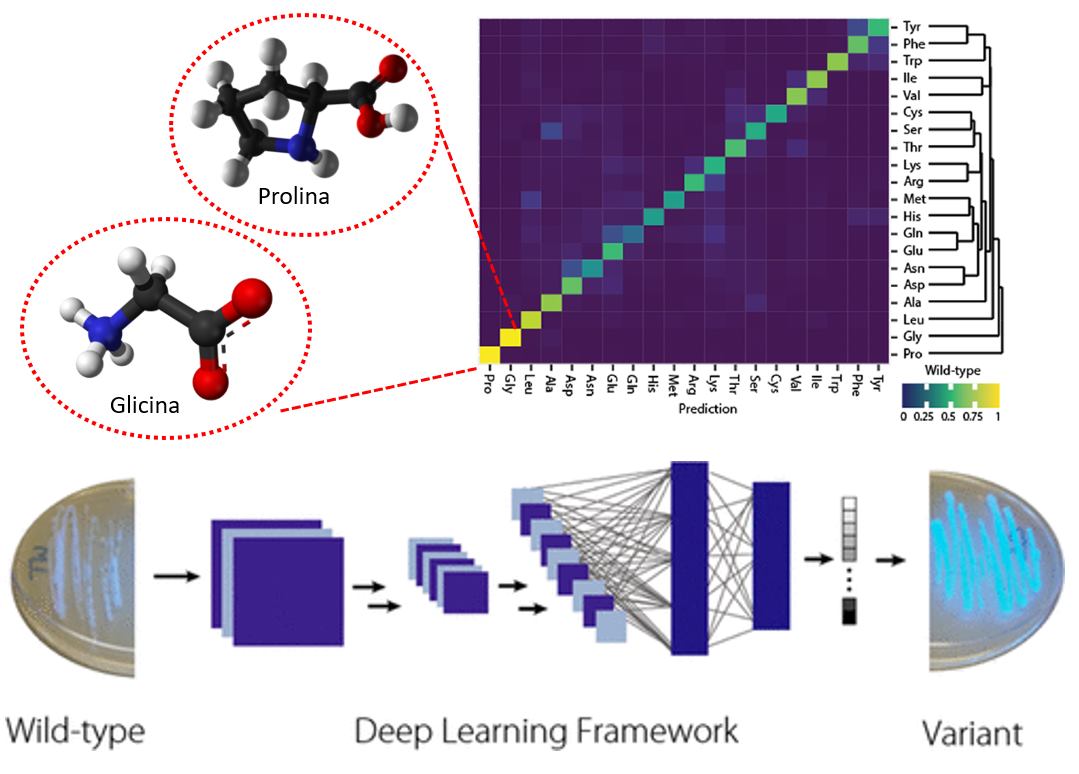
With this, the network learned to model the chemical microenvironments within proteins and recapitulate certain known properties of amino acids, such as Proline and Glycine, which have very characteristic and unique microenvironments that make them easier to distinguish from the rest (F3). With this network, they were able to identify amino acids prone to mutation and through experiments, they observed that each of these mutations has an individual and positive contribution to the function of different proteins they analyzed. However, if you combine these mutations, you can generate an additive effect on their function! For example, using all the mutations identified for a blue-fluorescent protein, they were able to improve its luminescence up to 6 times. In principle, a model like this allows one to operate with mutations as if they were sums to improve their functions.
Refs:
