Post 6: Thousands of new microbes 🌎
Published:
Not every day more than 6000 metagenomes get released all at once! Two days ago (09.11.2020), researchers from the “Joint Genome Institute” of the US Department of Energy published the results of the largest project to date in the field of metagenomics and prokaryotic genome reconstruction.
By analyzing ~4000 public metagenomes and adding ~6000 new metagenomes from different (sub)environments, they managed to capture new groups of bacteria and archaea. Compared to other similar projects, this project quadruples the diversity found compared to the previous one that had found the greatest biodiversity of groups [F1].
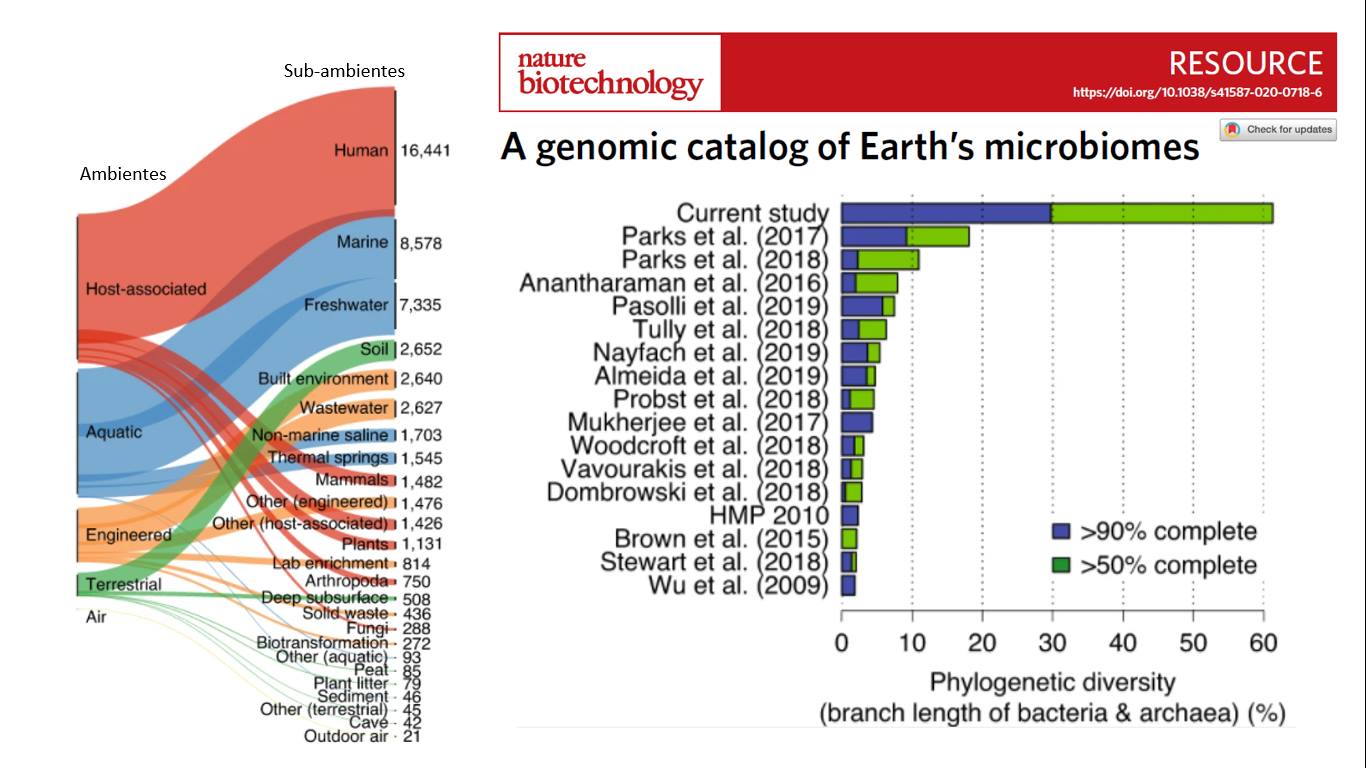
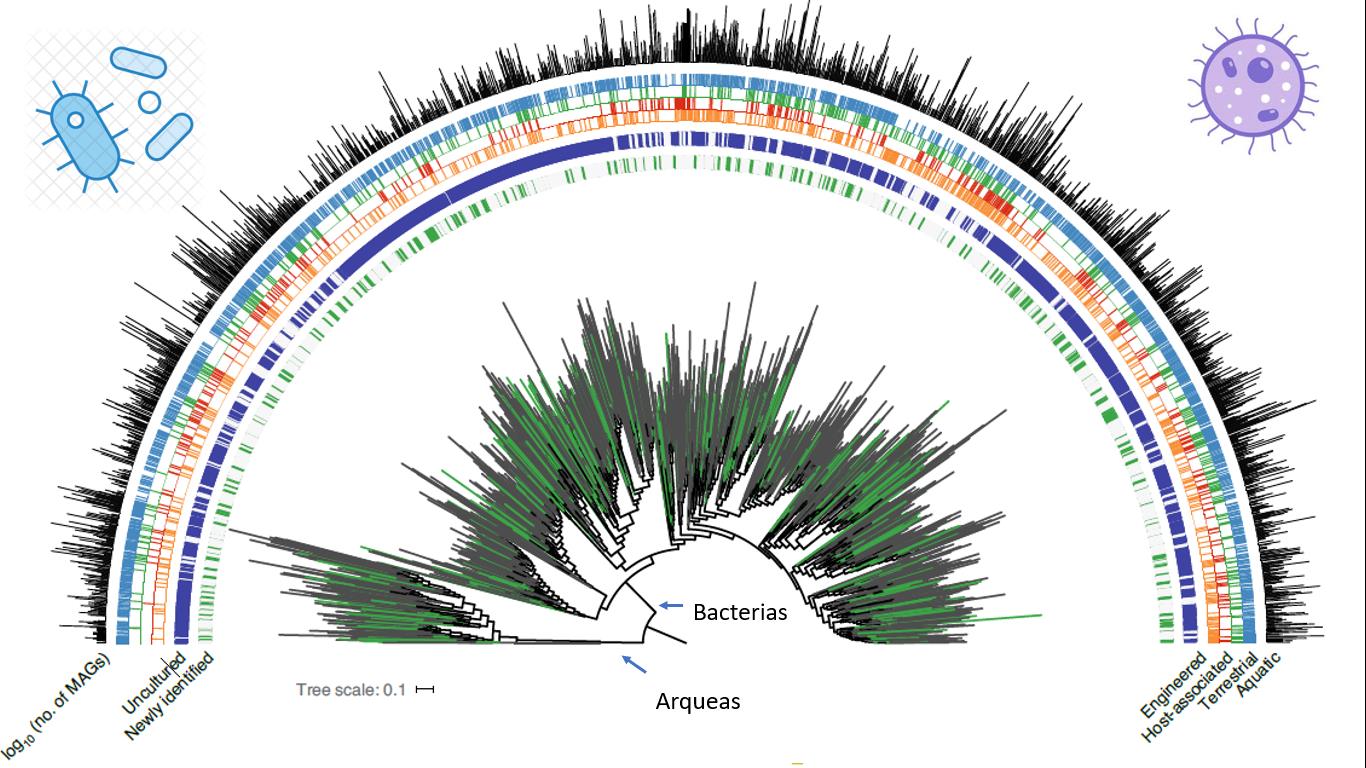
Specifically, they managed to reconstruct more than ~50,000 microbe genomes, of which ~12,000 are new groups: 16 phyla, 456 orders, 1,525 families, 5,463 new genera, are just some of the attributes of this project. Combining many public genomes with those they managed to reconstruct, they estimate that 75% of the prokaryotic biodiversity we know is represented only by genomes that we have not been able to cultivate in the laboratory [F2].
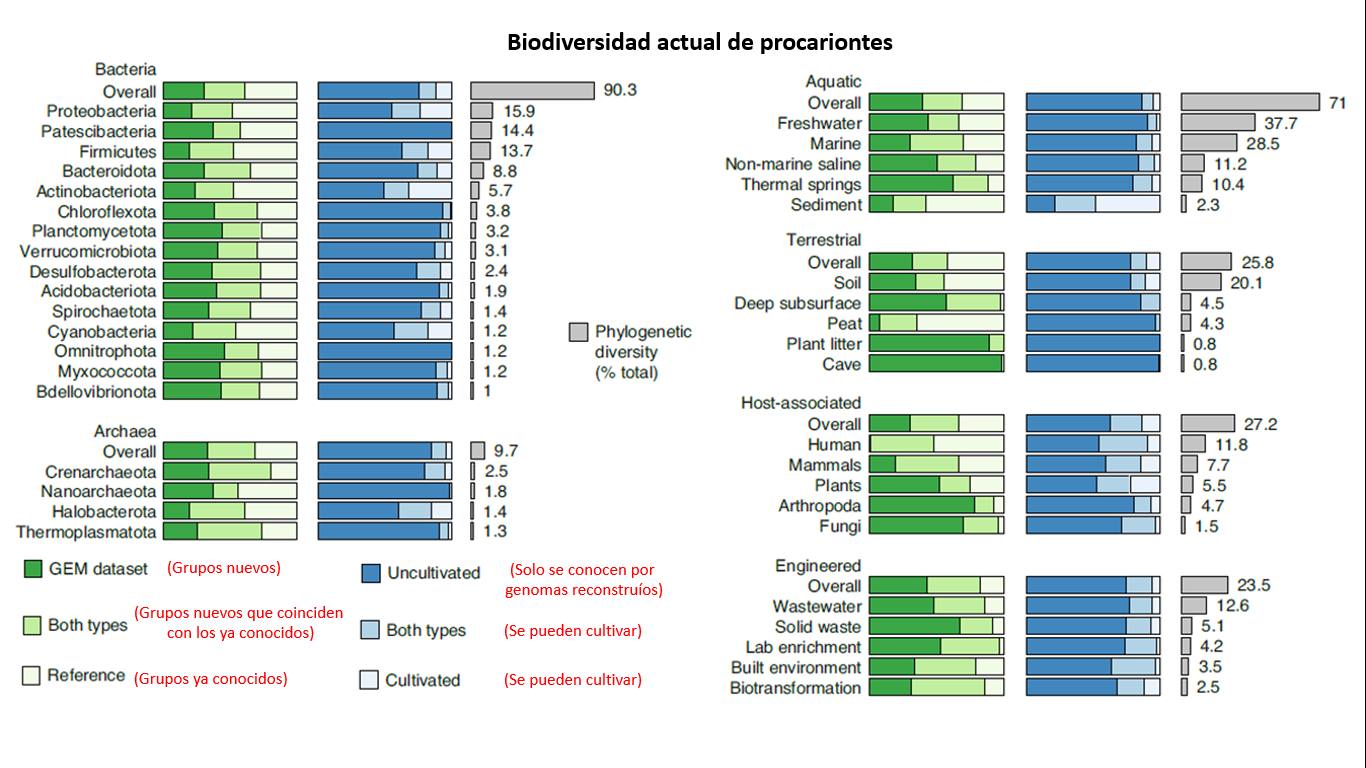
If we delve into analyzing the biodiversity in bacteria and archaea, we see that the former are the most diverse (in evolutionary terms), and within bacteria, the Proteobacteria, which are practically ubiquitous, are the most diverse, followed by the group discovered in 2014, the Patesibacteria, which have very small genomes, and finally the Firmicutes (about which I know nothing). If we analyze biodiversity at the environmental level, we can see that a large part resides in aquatic environments, where more than 80% of the groups are only known to us thanks to bioinformatics. Other environments that we practically only know through bioinformatics are caves, leaf litter, or peat [F3]. Interestingly, the only thing we can say we know relatively well is humans, where not many new groups were found, and ~50% of the microbes have already been cultivated.
In addition to everything, they also took on the task of analyzing the metabolic potential of all these genomes, as well as possible new natural products that have the potential to be exploited with biotechnology, and even more, to relate each of these prokaryotes to their respective viruses. But that’s another story.
Refs:
