Post 5: The human microbiome 👫
Published:
Alexandre Almeida is a scientist who, in addition to being the first author of the following research, helped me in my undergraduate thesis. For this reason, I dedicate this publication to him as a form of gratitude. He also taught me that no matter how busy you may be, there is always room to help someone who asks for it.
Microbes accompany us from birth to after death and are responsible for our health by helping us incorporate nutrients; for example, vitamin B12, essential for brain function, is exclusively produced by prokaryotes [F1]. Given their importance in our health, the “Human Microbiome Project” was initiated in 2008 to identify the microorganisms responsible for health and disease. The project progressed, and many bacteria were cultured and their genomes sequenced from body regions such as the mouth, vagina, skin, and intestines (from fecal matter), with the latter being the region of the body with the most bacteria.

With massive DNA sequencing (metagenomics, in this case), it has been possible to sequence the genomes of the microbes present in our intestines. In this sense, Alex’s work and his team show the great progress that has been made since 2008. By analyzing intestinal samples from 31 countries, they created the “Unified Collection of Human Gastrointestinal Genomes,” which comprises 4,644 species of prokaryotes (including 28 archaea) represented by 204,938 genomes. Additionally, they created the “Unified Catalog of Gastrointestinal Proteins,” which comprises ~15,000,000 protein clusters represented by 170,602,708 coding DNA sequences [F2].
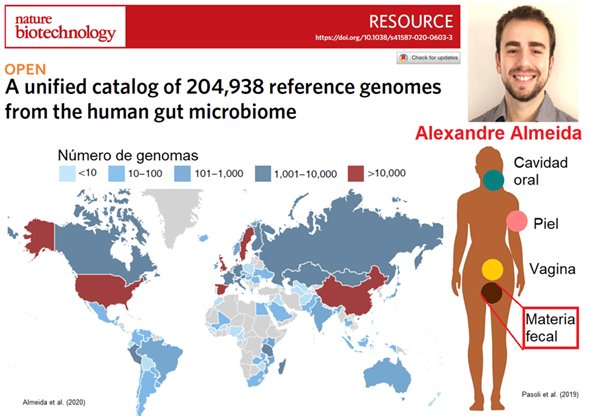
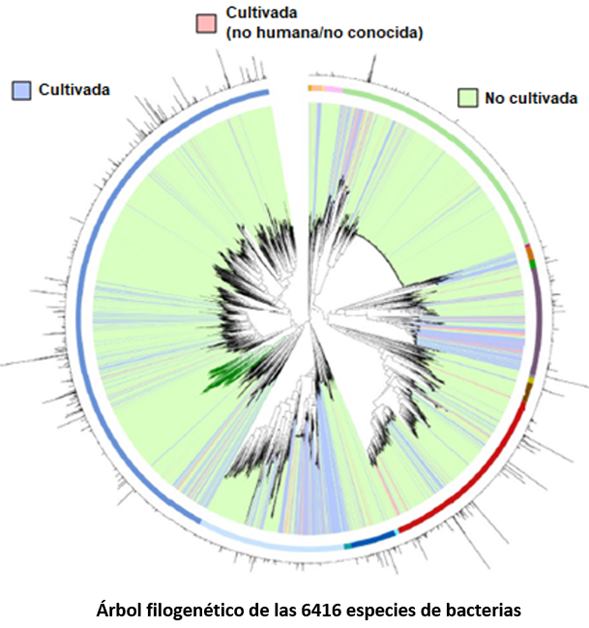
It is estimated that ~71% of the 4,644 species have not been successfully cultured in the laboratory, and many more species remain to be identified [F3]. However, their analysis is highly biased by the origin of the samples, as the vast majority come from people in the United States of America, China, and Europe. Nonetheless, the data suggest that the most abundant bacterial species in our intestines is Agathobacter rectalis, and Methanobrevibacter_A smithii is the most abundant among archaea. Additionally, it is suggested that there is a great variability at the strain level between continents and that curiously, 0.2% (34,070) of the protein clusters are of viral origin.
All the information and several more implementations are available to the community on the MGnify page. Regarding underrepresented regions like Latin America, hopefully, the MicrobioMEX initiative can be resumed soon [F1], which is currently stalled. Perhaps selling the idea in times of the 4T and EAB as a “Responsible social study of the microbiome of indigenous and urban communities in the Mexican Republic and its relationship with national corn and diabetes” could make it easier to obtain financing!
Refs:
