Post 11: Massive Microbial Phylogenies 🧬
Published:
Today is Microbiome Day! So, I’ll summarize my favorite article on microbial evolution.
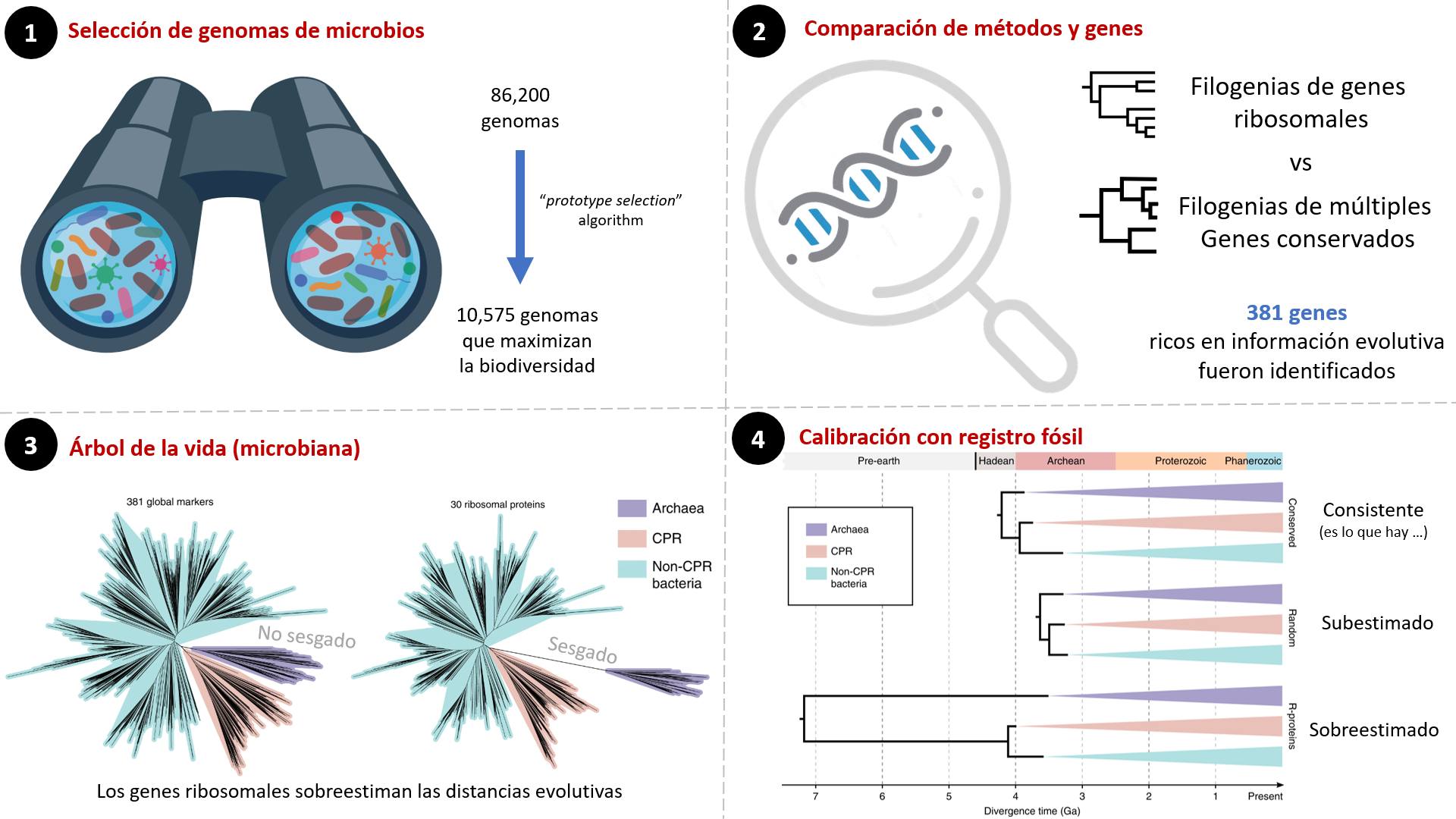
Qiyun’s team took on the task of analyzing the tree of life of microbes. This is not easy, as although we have many genomes, many are incomplete, and it is also not clear how to identify the best ones.
To identify representative genomes, they wrote an algorithm that seeks to maximize biodiversity based on genetic distances between microbes and found 10,575 representative genomes.
The next step is to identify genes rich in evolutionary information from these genomes. After several analyses, they identified 381 genes with this condition. And with these, they compared many methods of phylogenetic inference.
With these information-rich genes, they were able to reconstruct the tree of life of microbes. They observed that if we use only ribosomal proteins, the evolutionary relationships between bacteria and archaea are overestimated.
Finally, they added information from the fossil record to this tree of life to estimate the divergence times of many groups of microbes. They observed that if they use ribosomal proteins, these times are not consistent with estimates of the origin of life. If they take random genes, divergence times are underestimated. But if they take the 381 genes, they obtain very good consistency with what we know about biological and geological evolution.
It’s not an easy article to read, but it’s valuable. And it even comes with a website. https://biocore.github.io/wol/
Refs:
